
The secret of protoribosomes: Michal H. Kolář discusses ground-breaking research about how life evolved
Michal H. Kolář studied chemistry, focusing on theory and molecular modelling, and did his PhD at the Institute of Organic Chemistry and Biochemistry of the Academy of Sciences of the Czech Republic (IOCB Prague). He was a visiting researcher in South Korea and France. After postdoctoral work at the Forschungszentrum Jülich in Germany, where he had access to the (then) most powerful computer in Europe, and he also was a postdoctoral researcher at the Max Planck Institute for Biophysical Chemistry in Göttingen (Germany). There he began to investigate the synthesis of proteins and ribosomes, research he continues to this day. He was recently involved in a study of the protoribosome and results were published in Nucleic Acids Research (NAR).
What was the topic of the study for your collaboration with Klára Hlouchová’s team?
It was about the protoribosome, but before I get to that, I should explain what a ribosome is. A ribosome is a cellular machinery that is present, in many copies, in all living cells. It is responsible for protein synthesis. Ribosomes originated roughly 3.5 to 4 billion years ago and then continued to evolve into today’s form. We studied ribosome precursors that existed before life even appeared.
So what were the conditions on Earth at that time?
No one knows exactly. Since Alexander Oparin published The Origin of Life, there has been talk about a so-called primordial soup, an alleged mixture of short biopolymers (proteins and nucleic acids), their building blocks, and similar small molecules that were likely created from simple molecules such as ammonia or methane. We think these short polymers gradually began to associate together in water, to varying degrees. Some acquired catalytic abilities and thus, simple reaction networks began to emerge. From these primordial biomolecules and biomolecular complexes, something larger and more “viable” was “selected”, not in the Darwinian sense of natural selection but rather a kind of biophysical optimization of reaction processes.
And at one point, the ribosome probably appeared in the primordial soup.
More or less, yes. But it didn’t appear out of nowhere. Rather, ribosome precursors grew gradually. Ribosomes are relatively large particles of about 25 nanometres in diameter. In bacteria, it has about 50 proteins and 3 strands of ribosomal RNA. The ribosome size gradually increased during biophysical optimization and later evolution, which indicates that the interior of a ribosome is evolutionary older than its surface. We studied the biomolecular precursors of the ribosome, the so-called protoribosomes, which are much smaller than the modern ribosome but which show catalytic activity. Recently, 2009 Nobel Prize laureate in chemistry, Ada Yonath, and her research team, showed that protoribosomes are able to catalyse the formation of peptide bonds. So we focused our own study on the circumstances under which protoribosome is formed.
So what did you discover?
We studied two protoribosomes, a large one and a small one (both still much smaller than a ribosome). It turns out that when RNA associates together with short peptides, it becomes more stable. This is probably one of the mechanisms by which the protoribosomes would be selected as efficient catalysts. In the study, we showed that a protoribosome composed of RNA and short peptides is more resistant to UV radiation and enzymes than a protoribosome without peptides.
So what makes this study significant?
It combines various experimental techniques with computer simulations, providing new insight into how a complex network of chemical reactions led to biology, i.e. life, though we still don’t know many things. In this study, we described a mechanism that may have contributed to the emergence of the modern ribosome.
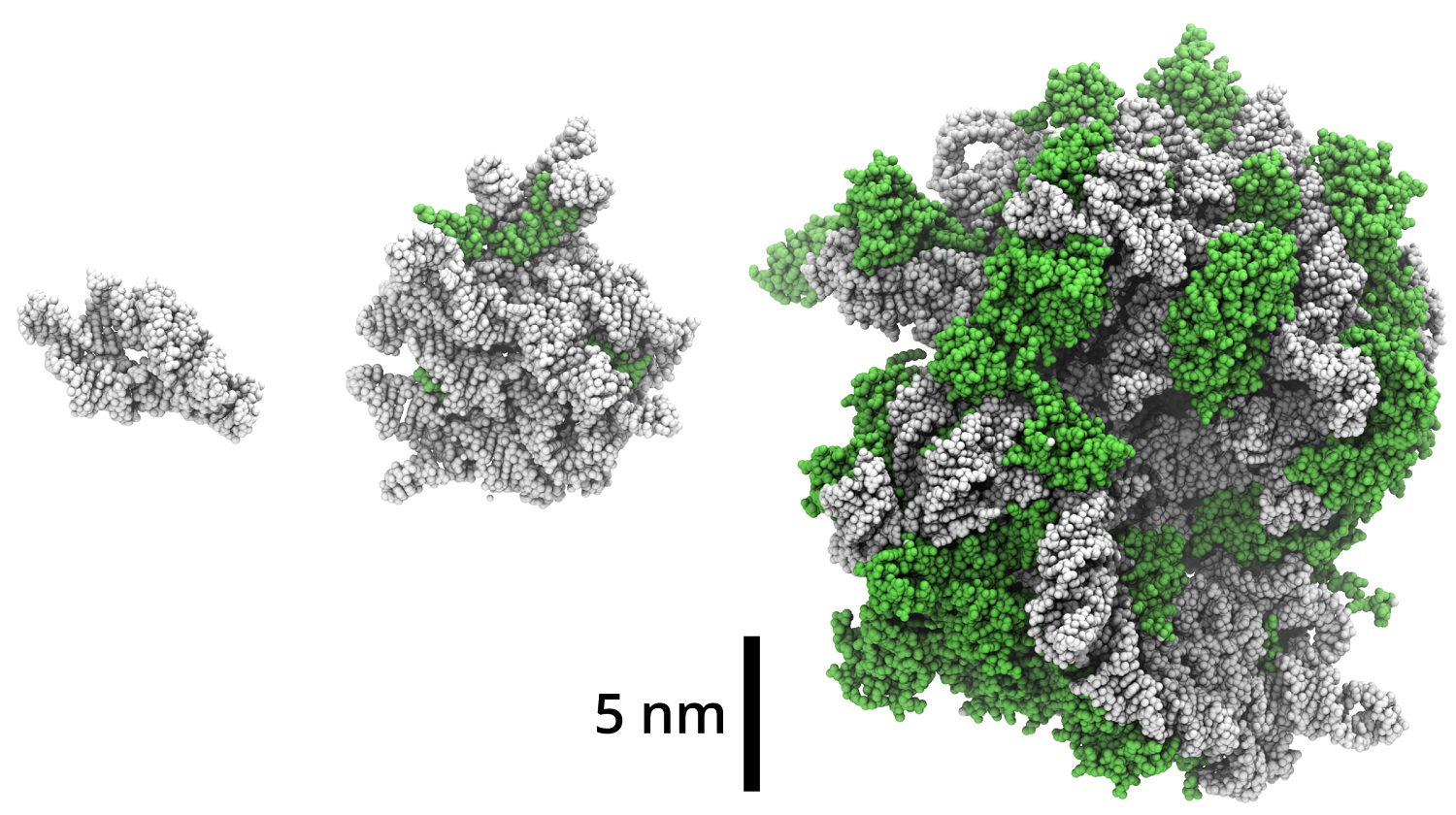
Figure: Protoribosomes (left and middle) in this study in the context of the current bacterial ribosome (right). RNA is gray, proteins are green. The smaller of the studied protoribosomes is consisted only of RNA; while several short proteins are attached to the larger protoribosome.
Who were UCT Prague participants in this study?
It was very collaborative, with researchers from seven institutions in the Czech Republic, Italy, and Japan. Klára Hlouchová from Charles University led the team. She easily connects people in meaningful and effective collaborative efforts. Martin Mašek, at that time my undergrad student, and I were the UCT Prague representatives. Martin did his bachelor’s thesis on this topic and he’s now continuing his master’s studies. Much credit goes to Martin. I remember how he started working on this topic during an Erasmus internship in Portugal. We dealt with everything only on Zoom for the first half year of the project.
How many people worked on the study, and for how long?
I confess that I don’t really know how long the experiments were going on. We were invited to join while experiments were already going on and we collaborated on the study about a year. The NAR publication has 10 co-authors.
What was your role?
We contributed with our expertise on ribosomes and computer simulations, supplementing the experimental part of the study with the context of the modern ribosome and a molecular perspective. Martin and I were in charge for simulations and their evaluation.
What did your simulations show?
The main conclusion from the simulations we performed is that the larger protoribosome was structurally more stable and less flexible than the smaller one. The proteins we studied bound specifically to the larger one. It means that there is a specific place on protoribosomal RNA where the proteins bind. On the other hand, in the smaller protoribosome, proteins bind in a less specific way: the protein and RNA are still in contact, but the protein somehow “crawls” along the RNA.
What equipment did you use for the simulations?
We used the largest Czech supercomputer from IT4Innovations in Ostrava, specifically the Karolina supercomputer. I cannot praise IT4I enough for our cooperation with IT4I for their fantastic management of their supercomputing systems.
What impact does this study have on the perception of the origin of life and evolution?
The so-called RNA world hypothesis dominates academic discourse on this topic. According to this hypothesis, various self-replicating RNA molecules existed prior to emergence of proteins and DNA, the building blocks of life that we see now. The function of RNA, according to this hypothesis, was to provide catalysis and information storage. Our research supports an alternative hypothesis, according to which peptides and short RNAs coexisted and jointly participated in the biophysical optimization of reaction networks and the formation of longer biopolymers.
What challenges did you face during this study?
The biggest challenge was to putting everything together into a coherent story. Klára takes the main credit for that.
And is there anything that pleasantly surprised you?
I like ribosomes. They are large enough that you can look at different segments and find various questions that have yet to be answered. Yet ribosomes put large demands on simulations because of their large size and complex chemical composition. With the protoribosome, all of sudden, we got a much smaller particle in our sights, and our methodology was perfectly suited to studying it. I really enjoy the new kinds of questions we are able to address with simulations.
What are you planning next? Are you going to follow up on this study with the same team?
Yes, we have many plans. We would like to study even smaller protoribosomes, how they are affected by their environment and by their clustering. This spring, we applied for funding from The Czech Science Foundation, so we are waiting to hear from them. Personally, I’d would be very glad if simulations, my area, would play a more prominent role in the forthcoming projects.
Reference
The interplay between peptides and RNA is critical for protoribosome compartmentalization and stability (Nucleic Acids Research, 2024)







